Project:
| Project name: | "Development of cancer forecasting infrastructure based on genome and health data" |
| Objective of the specific aid / title of the measure: | 1.2.1. Specific support objective “Increase private sector investment in R&D” 1.2.1.2. Measure “Support for the Improvement of the Technology Transfer System” |
Project applicant: (research organization) | University of Latvia |
Cooperation agreement no..* | KC-L-2017/5 |
| Project identification no. | KC-PI-2017/102 |
Date: | 30.07.2018 |
* Agreement concluded between the research organization and LIAA on receiving support within the framework of the commercialization fund.
Description of the problem
Today, there is a lot of data that is created, processed and analyzed on a global scale. The steady increase in data covers almost all sectors and healthcare is no exception. By reducing the cost and time of human genome sequencing, as well as increasing the number of medical examinations and diagnostic procedures, healthcare as an industry provides a solid basis for data-driven decision-making not only for personalized treatment of traditional chronic diseases but also for personalized prevention.
Much of the medical data is already available in the healthcare ecosystem, but current technological capacity to process large amounts of structured and unstructured data from heterogeneous resources stored in different forms to predict disease likelihood is limited. This, in turn, reduces the ability to provide an individual and highly tailored response to genetic and diagnostic findings, thus reducing the wide range of effective prevention options for chronic diseases (such as cancer) and personalized treatment.
Risinājums

The video cannot be viewed because you have not selected functional cookies.
To view the video, please enable functional cookies in your cookie settings.

The video cannot be viewed because you have not selected functional cookies.
To view the video, please enable functional cookies in your cookie settings.
Latvijas Universitāte (LU) ir konceptualizējusi datu analīzes platformu, kas būtu īpaši pielāgota gēnu un veselības datu analīzei, un pēc tam izveidotu uz datiem balstītu infrastruktūru (datu ezeru), kas rezultātā sekmētu efektīvāku lēmumu pieņemšanu un pielāgojamu un rentablu veselības aprūpes sistēmu.
Analītiskās infrastruktūras izveidošanas sākotnējais izmantošanas veids ir plaušu vēzis, jo jau ir pietiekami daudz ģenētisko un epidemioloģisko datu, lai izveidotu un pārbaudītu platformas analītiskās spējas, taču tiek uzskatīts, ka šī platforma tiek attīstīta tā, lai to izmantotu lietošana arī citiem medicīnas lietojumiem.
Analītisko platformu izmantos gan zinātnieki kā tīru analītisku medicīnas datu instrumentu, gan veselības aprūpes politikas plānotāji un medicīnas iestādes, lai izstrādātu uz datiem balstītas slimību ārstēšanas un profilakses plānus.
Application of technology
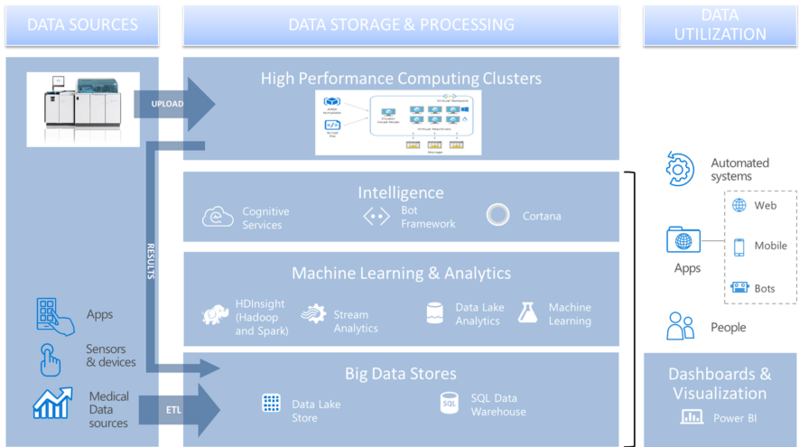
The aim of the project is to develop the necessary IT support infrastructure for the collection of gene and health data, with the ability to integrate and analyze them.
Estimates will be health-related, but specific methods are currently unknown as they will depend on specific project requirements (common estimates such as descriptive statistics will be available, but specific analytical circuits will be developed for the pilot project, making them adaptable where possible). ).
The developed architecture will provide researchers with a single workplace with a shared database, which would reduce the number of redundant tests used in different projects. Genome sequencing results, as well as the results of other tests and patient descriptive data, can be reused in many projects and analyzed from different perspectives, resulting in higher returns on initial investment.
This project will provide IT solutions for the validation of architectures in the relevant environment (TRL 5) and interface testing to facilitate the uploading, consolidation, processing, analysis and use of large-scale structured and unstructured genome and health data from a variety of sources. This means that the architecture and infrastructure will be ready for data analysis and analysis in different projects, and the infrastructure specific to this project will be tested in the specific project (lung cancer) and thus adapted for future use in other projects.
The architecture of these solutions can be used in both research and precision medicine and will support the following five interrelated applications:
- Data mining;
- Data processing and anonymisation;
- Data analysis;
- Providing data in research and precision medicine;
- Analysis of user data in the interface and infrastructure environment.
As a result of the project, a data-based infrastructure (data lake) will be developed, which will provide full-fledged data and interface solutions for more efficient treatment decisions, thus promoting the development of a more cost-effective health care system. The five use cases described above will be tested on Latvian genome and health care data, which will allow the development of new methods for risk assessment, diagnosis, prognosis and determination of therapeutic efficacy in the treatment of lung cancer.
The beginning of the project - a trial based on medical data of patients living in Latvia, initially lung cancer and general health data, later - data characteristic of other diagnoses.
Team
This project will be implemented in cooperation by the Faculty of Computer Science, the Faculty of Medicine and the Faculty of Business, Management and Economics of the University of Latvia. The project team is led by Signe Bāliņa, a leading IT researcher with more than 20 years of scientific experience, many publications, as well as more than 10 years of experience in managing a software development company, where she was directly responsible for the development of various IT products.

- LU Inovāciju granti studentiem
- Projekts “Virtuālās prakses Baltijā”
- DIGITEKA
- Collaboration with Microsoft IoT Insider Labs
- Cooperation with a venture capital fund
- Unmanned car test track
- Development of cancer forecasting infrastructure based on genome and health data
- Digital platform for research project and competence management
- The large data is based on a healthy human intestinal metagenoma model and its applications
